This strategy led to the amplification of three different segments of the receptor codon. Fragment I, extending from position 1242 to position 1919, included the found point mutations at position 1515 and 1786. Fragment II, from position 1721 to position 2639, contained the single base substitution at position 2316. The location of these fragments was chosen due to the distribution of the restriction sites of the restriction endonucleases in question (see section usedRE).
The third fragment, reaching from position 1284 to position 1466, included the
amino acid changing point mutation of position 1309 (GGA ![]() AGA, see
section results). As this mutation does not alter any restriction site,
a BfaI site was introduced by altering the
last but one nucleotide of the 5' oligonucleotide primer of the fragment. By
this means the nucleotide change at position 1309 resulted in the creation of a
restriction site for the AGA allele (coding arginine). The introduction of the
BfaI site was constructed because it required the change of one
nucleotide only. The used PCR primers and size of the amplified fragments are
shown in table tab:RFLPprimers.
AGA, see
section results). As this mutation does not alter any restriction site,
a BfaI site was introduced by altering the
last but one nucleotide of the 5' oligonucleotide primer of the fragment. By
this means the nucleotide change at position 1309 resulted in the creation of a
restriction site for the AGA allele (coding arginine). The introduction of the
BfaI site was constructed because it required the change of one
nucleotide only. The used PCR primers and size of the amplified fragments are
shown in table tab:RFLPprimers.
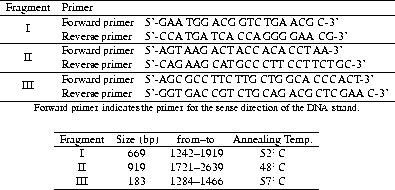
Table 4.4: Nucleotide sequence of DNA primers used for PCR amplification for RFLP
of the ![]() adrenoceptor codon (above) and their characteristics (below)
adrenoceptor codon (above) and their characteristics (below)