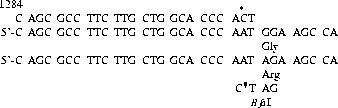
The restriction endonuclease used for the third fragment resulted from the mismatch introduced into the 5' oligonucleotide primer as shown below:
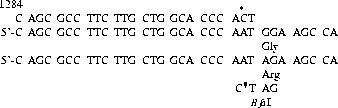
This construct leads to an uncut fragment with a length of 183 bp for the glycine allele (GGA) and to two fragments, 159 and 24 bp in length, for the arginine allele. Therefore, in a 2.5% agarose gel, you can expect a 183 bp band for a homozygous GGA amplificate, the cut 159 bp band for a homozygous AGA amplificate, and both bands in heterozygous amplificates (see figure fig:ag_bfa1309).
The first fragment includes changed restriction sites for the mutations at
position 1515 and 1786. The point mutation located at position 1515 destroys a
BfaI restriction site. The mutation at
position 1786 introduces five new restriction sites (see table
tab:1786re). KpnI is the only rare
cutting one as the complete ![]() adrenergic receptor structure lacks this
specific site type.
adrenergic receptor structure lacks this
specific site type.
The single base change at position 2316 (see section results) destroys a BsrDI site . The used fragment was selected to include another BsrDI cleavage site for reference. The used restriction endonucleases and the resistant fragments are summarized in table tab:REs.
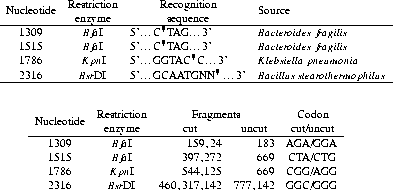
Table 4.5: RFLP fragments and used restriction endonucleases