The enzymatic method is based on the ability of a DNA polymerase (Taq polymerase ) to extend a primer, hybridized to the template to be sequenced, until a chain-terminating nucleotide is incorporated. Each sequence determination is carried out as a set of four separate reactions, each of which contains all four dNTPs supplemented with a limiting amount of one dideoxyribonucleoside triphosphate (ddNTP). Because the ddNTP lacks the necessary 3'-hydroxyl group required for chain elongation, the growing oligonucleotide is terminated selectively at G, A, T, or C, depending on the respective dideoxy analog in the reaction. The relative concentrations of dNTPs and ddNTPs can be adjusted to give a nested set of terminal chains several hundred bases in length. For a detailed summary of this reaction, see also section seq.
Incorporating a radiolabel somewhere in the oligonucleotide chain permits the
visualization of the sequencing products by autoradiography . Two basic
radiolabeling protocols can be utilized to detect the reaction products. The
incorporation labeling method, developed by TABOR and
RICHARDSON[121], separates the sequencing reaction into a
labeling step and an extension/termination step. In the first step, the primer
is extended a short distance using limiting concentrations of the dNTPs and a
single radiolabeled dNTP. In the second step, the extended primers are
further extended in the presence of both dd- and dNTPs. Using the direct
labeling method, a label is directly attached to the end of the
primer[122, 123, 124]. The oligonucleotide is 5' end-labeled using T4
Polynucleotide Kinase and [ ![]() -
- ![]() P]dATP. The subsequent
extension/labeling reaction is not limiting for one of the dNTPs.
For this study, the direct incorporation protocol with [
P]dATP. The subsequent
extension/labeling reaction is not limiting for one of the dNTPs.
For this study, the direct incorporation protocol with [ ![]() -
- ![]() S]dATP
was used[125].
S]dATP
was used[125].
[ ![]() -
- ![]() S]dATP (see figure fig:satp) was preferred to [
S]dATP (see figure fig:satp) was preferred to [ ![]() -
- ![]() P]dATP for several
reasons. The strong particles emitted by
P]dATP for several
reasons. The strong particles emitted by ![]() P create two problems. First,
because of scattering, the bands on the autoradiograph are far larger than the
bands of DNA in the gel. This affects the ability to read the sequence
correctly and limits the number of nucleotides that can be read from a single
gel. Second, the decay of
P create two problems. First,
because of scattering, the bands on the autoradiograph are far larger than the
bands of DNA in the gel. This affects the ability to read the sequence
correctly and limits the number of nucleotides that can be read from a single
gel. Second, the decay of ![]() P causes radiolysis of the DNA in the
sample. Sequencing reactions radiolabeled with
P causes radiolysis of the DNA in the
sample. Sequencing reactions radiolabeled with ![]() P can therefore be stored
for only 1 or 2 days before the DNA is so badly damaged that it generates
indecipherable sequencing gels. The use of [
P can therefore be stored
for only 1 or 2 days before the DNA is so badly damaged that it generates
indecipherable sequencing gels. The use of [ ![]() S]dATP [125]
greatly alleviates both problems. Because of the weaker
S]dATP [125]
greatly alleviates both problems. Because of the weaker ![]() particles
produced by decay of
particles
produced by decay of ![]() S, there is little or no loss of resolution between
the gel and the autoradiograph. Furthermore, the lower energy produces less
radiolysis, allowing sequencing reactions to be stored for up to 3 weeks at
-20psy176 C without noticeable loss of resolution. Though, longer exposure
times of the autoradiograph are necessary.
S, there is little or no loss of resolution between
the gel and the autoradiograph. Furthermore, the lower energy produces less
radiolysis, allowing sequencing reactions to be stored for up to 3 weeks at
-20psy176 C without noticeable loss of resolution. Though, longer exposure
times of the autoradiograph are necessary.
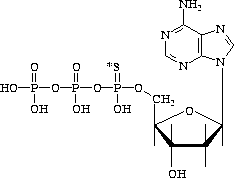
Figure 3.4: [ ![]() S]-Deoxyadenosine 5'-[
S]-Deoxyadenosine 5'-[ ![]() -thio]triphosphate
-thio]triphosphate
The primer was annealed with
the dsDNA plasmid template in a molar ratio of approximately 1:1. The 18 ![]() l
sample from the alkali denaturation step (see section seqdenat) was
splitted into two 9
l
sample from the alkali denaturation step (see section seqdenat) was
splitted into two 9 ![]() l volumes for two sequencing reactions with different
primers. 2
l volumes for two sequencing reactions with different
primers. 2 ![]() l primer
l primer![]() (approx. 2 pmol), 2
(approx. 2 pmol), 2 ![]() l extension/labeling mix, 5
l extension/labeling mix, 5
![]() l Taq polymerase 5
l Taq polymerase 5 ![]() buffer and water to 25
buffer and water to 25 ![]() l were
added. This reaction was incubated at 37psy176 C for 10 minutes.
l were
added. This reaction was incubated at 37psy176 C for 10 minutes.
2 ![]() l of [
l of [ ![]() -
- ![]() S]dATP (1,000 Ci/mmol,
approximately 10
S]dATP (1,000 Ci/mmol,
approximately 10 ![]() Ci/
Ci/ ![]() l, DuPont NEN) were added to the annealed
primer/template mixture. 1.5
l, DuPont NEN) were added to the annealed
primer/template mixture. 1.5 ![]() l sequencing grade
Taq polymerase (7.5 U) were added
to the reaction and shortly centrifuged, and incubated at 37 psy176 C for 5
minutes. The extension/labeling reaction was carried out at 37psy176 C rather
than 70psy176 C to slow down the incorporation rate of Taq
polymerase and thereby limit the number of bases incorporated in this
step
l sequencing grade
Taq polymerase (7.5 U) were added
to the reaction and shortly centrifuged, and incubated at 37 psy176 C for 5
minutes. The extension/labeling reaction was carried out at 37psy176 C rather
than 70psy176 C to slow down the incorporation rate of Taq
polymerase and thereby limit the number of bases incorporated in this
step![]() . The incorporation of nucleotides is also limited by the
limiting concentration of nucleotides present in the extension/labeling mix.
. The incorporation of nucleotides is also limited by the
limiting concentration of nucleotides present in the extension/labeling mix.
For each set of the sequencing reactions, four microcentrifuge tubes (0.45 ml)
are labeled (A, C, G, and T) and 1 ![]() l of the appropriate d/ddNTP mix is
added to each tube. This nucleotide mix has a limiting amount of the specific
base (e.g., dGTP), which is also included as dideoxy nucleotide (ddGTP). The
loaded tubes are stored at 4psy176 C until just before completion of the
extension/labeling step.
When the extension/labeling reaction is complete, 6
l of the appropriate d/ddNTP mix is
added to each tube. This nucleotide mix has a limiting amount of the specific
base (e.g., dGTP), which is also included as dideoxy nucleotide (ddGTP). The
loaded tubes are stored at 4psy176 C until just before completion of the
extension/labeling step.
When the extension/labeling reaction is complete, 6 ![]() l are aliquoted to
each tube containing the d/ddNTP mix. They are briefly mixed by pipetting up
and down, and spun to ensure that no liquid is left on the tube walls. The
reaction is incubated at 70psy176 C for 5 to 10 minutes, and stopped by adding
4
l are aliquoted to
each tube containing the d/ddNTP mix. They are briefly mixed by pipetting up
and down, and spun to ensure that no liquid is left on the tube walls. The
reaction is incubated at 70psy176 C for 5 to 10 minutes, and stopped by adding
4 ![]() l of stop solution (95% deionized formamide, 20 mM EDTA, 0.05%
bromophenol blue and 0.05% xylene
cyanol) to each tube. [
l of stop solution (95% deionized formamide, 20 mM EDTA, 0.05%
bromophenol blue and 0.05% xylene
cyanol) to each tube. [ ![]() -
- ![]() S]dATP labeled reactions can be stored
at -20psy176 C for 2-4 weeks.
S]dATP labeled reactions can be stored
at -20psy176 C for 2-4 weeks.